Microbial vertical diversity in core sediments and its response to environmental factors near the hydrothermal field of the southern Okinawa Trough
-
摘要:
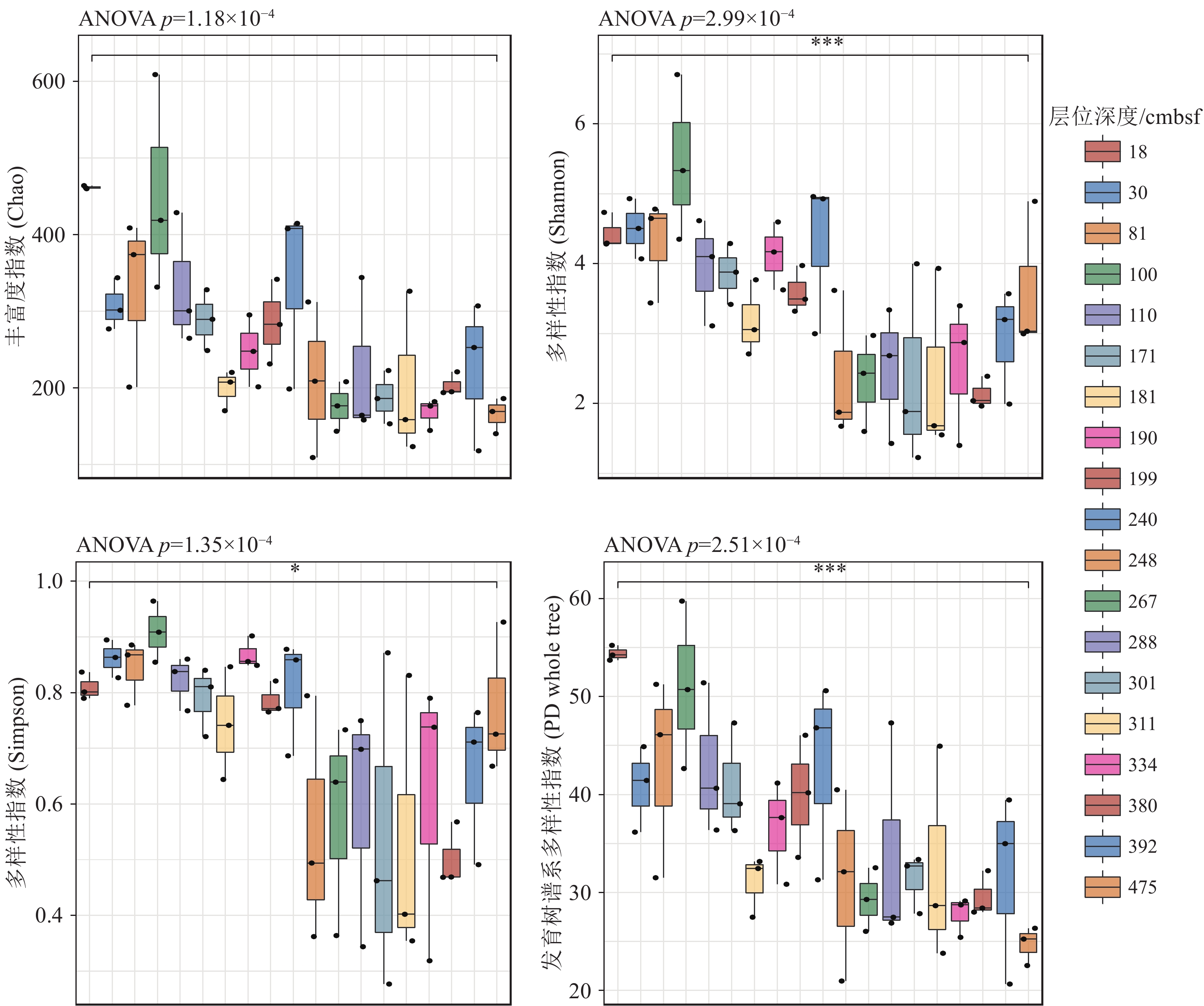
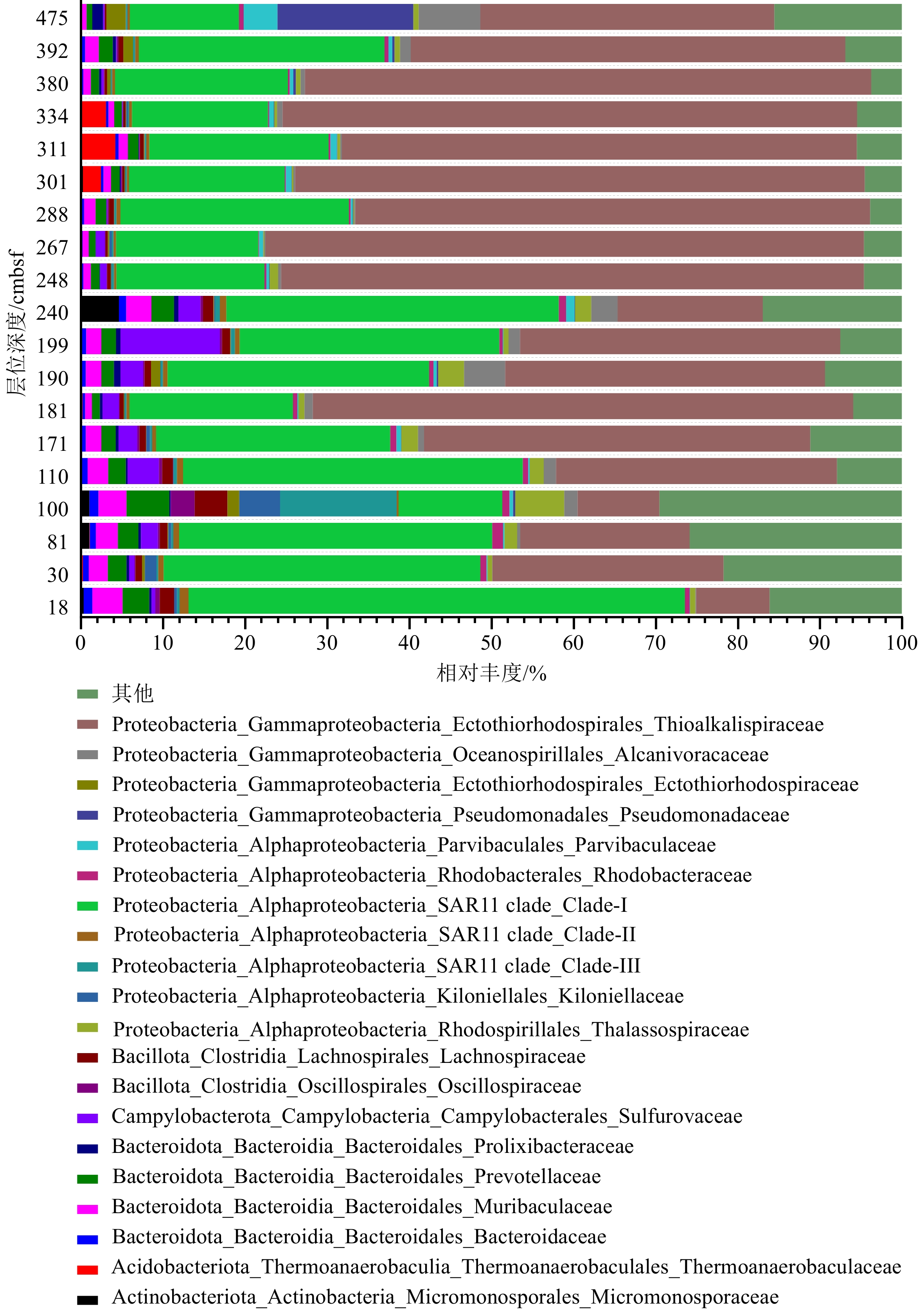
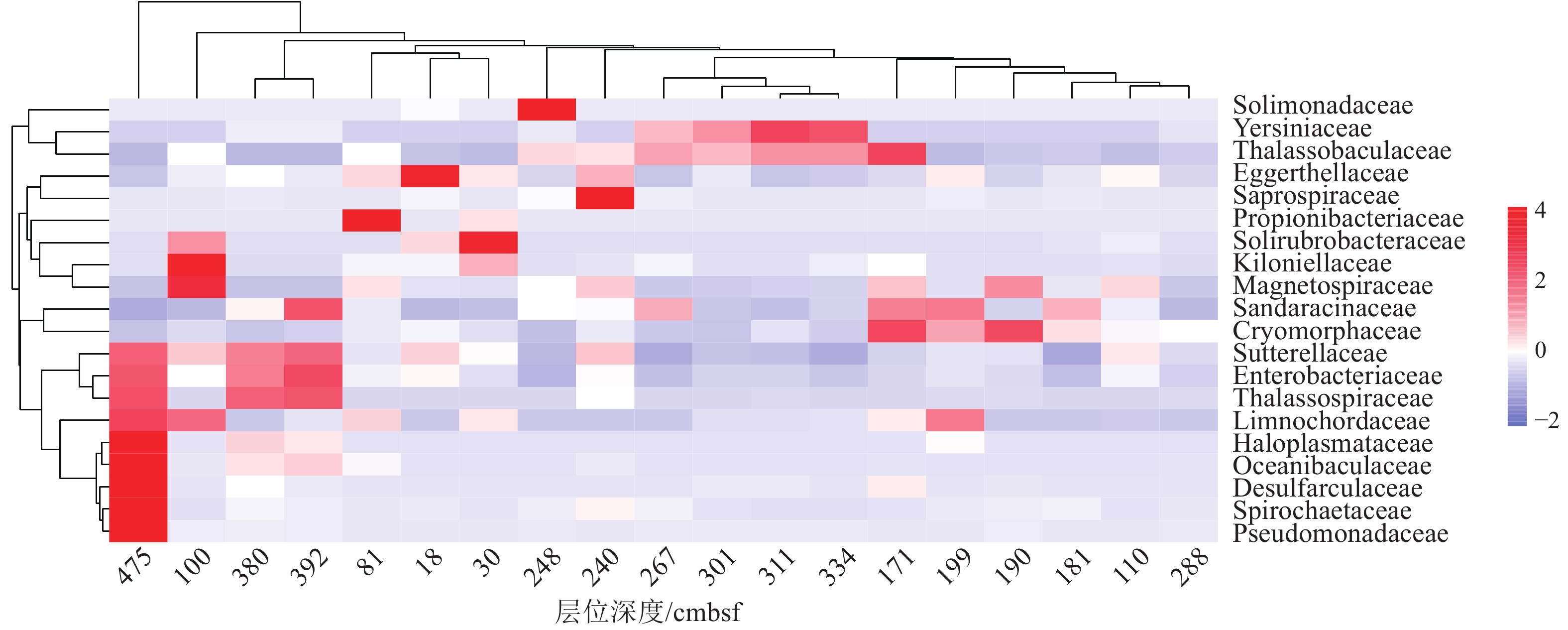
近年来,海底热液环境中的微生物及其环境适应机制已经成为海洋科学研究的热点。目前,相关的研究主要集中在表层沉积物及微生物的水平分布多样性方面,而对柱状沉积物中微生物垂直分布多样性研究却很少。本文基于西太平洋冲绳海槽南部热液区附近S2站位的柱状沉积物样品,通过对其不同层位的样品进行分离培养和16S rRNA基因高通量测序,揭示了样品中可培养微生物和总体微生物的垂直群落分布特征,同时结合对样品主量元素、微量元素、碳氮含量等指标的评估和冗余分析等统计学方法,讨论了微生物群落结构及其对环境因子的响应。研究发现该位点的柱状沉积物有机质含量较为贫乏,存在两个富含Cu-Zn-Pb的层;各个层位的沉积物中微生物类群均以变形菌为主要类群,同时表层沉积物表现出更高的微生物多样性。此外研究还表明柱状沉积物中有机碳含量与其微生物的群落组成有着更为密切的关系。总之, 本研究的结果和获得的菌种资源为进一步深入研究海底热液环境中微生物参与元素地球化学循环的过程提供了一定的基础。
Abstract:In recent years, microbe and its adaptation mechanism in submarine hydrothermal environment have become the focus of marine science research. At present, relevant researches focus on the horizontal distribution diversity of surface sediments and microorganisms, and only few researches on the vertical distribution diversity of microorganisms in columnar sediments. Based on the columnar sediment samples from the S2 station in the southern hydrothermal area of the Okinawa Trough in the western Pacific Ocean, we revealed the vertical community distribution characteristics of culturable microorganisms and the overall microorganisms in the samples through isolation, culture, and high-throughput sequencing of 16S rRNA gene from the samples at different levels. At the same time, the microbial community structure and its response to environmental factors were discussed by using statistical methods such as the evaluation of major elements, trace elements, carbon and nitrogen contents, and redundancy analysis. Results show that the organic matter content of the core sediment at this site is relatively poor, and Cu-Zn-Pb is rich in two layers; the microbial community of each layer is composed of mainly Proteobacteria, while the surface sediments exhibit higher microbial diversity. Meanwhile, it indicated a closer relationship between the organic carbon content of core sediments and the composition of their microbial communities. This study obtained bacterial strain resources and provided a basis for further research into microbial participation in the geochemical cycle of elements in submarine hydrothermal environment.
-
-
表 1 本研究使用样品的地球化学组成
Table 1. Geochemical compositions of the samples used in this study
样品深
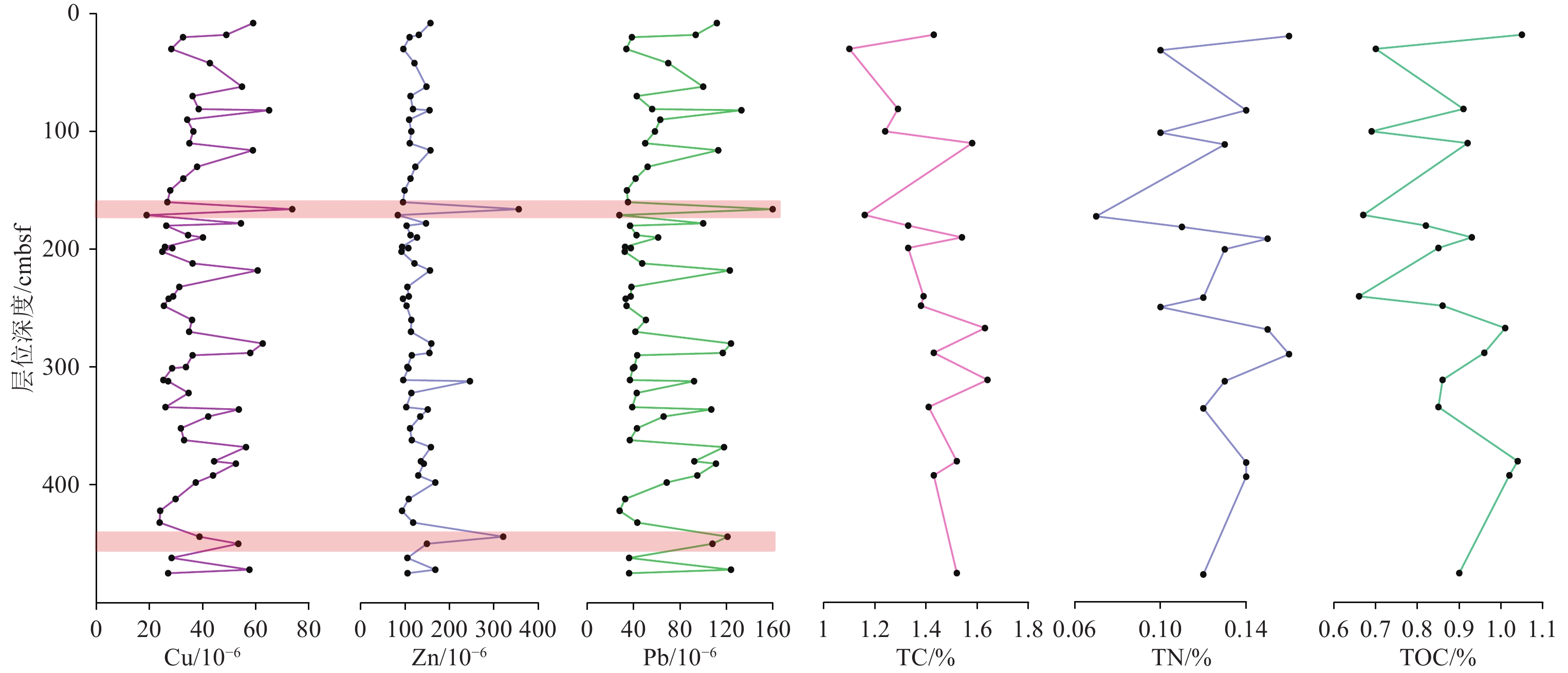
度/cmbsf18 30 81 100 110 171 181 190 199 240 248 267 288 301 311 334 380 392 475 Na2O/% 2.03 1.98 1.81 1.9 1.86 1.98 1.8 1.84 1.84 1.79 1.79 1.78 1.95 1.89 1.96 1.83 1.89 2.02 1.88 MgO/% 2.64 2.3 2.49 2.5 2.59 2.03 2.43 2.65 2.58 2.4 2.38 2.41 2.71 2.58 2.48 2.39 2.54 2.57 2.49 Al2O3/% 16.32 15.39 16.33 15.77 15.23 14.25 16.11 16.65 16.31 16.37 16.2 16.41 16.13 15.92 15.31 15.98 15.96 15.9 16.37 SiO2/% 58.88 60.43 59.27 59.7 59.17 62.59 59.43 58.05 57.99 59.26 59.53 59.27 59.12 58.48 58.99 59.51 59.62 58.96 58.55 Fe2O3/% 6.43 5.85 6.43 6.03 5.98 5.09 6.21 6.41 6.43 6.31 6.18 6.23 6.32 6.23 5.8 6.14 6.26 6.24 6.26 K2O/% 3.26 2.97 3.27 3.11 3.09 2.76 3.26 3.46 3.35 3.29 3.23 3.26 3.28 3.23 3.04 3.21 3.24 3.17 3.28 CaO/% 2.21 2.67 2.4 2.66 3.52 2.92 2.6 2.66 2.92 2.55 2.6 2.49 2.41 3.13 3.46 2.68 2.52 2.62 2.75 P/10−6 706.1 714.3 684.3 674 632.5 623.6 631.4 607.2 635.4 642 649.2 689.1 646.6 602.5 653.1 620.1 623 899 618.5 S/10−6 761.1 859.7 649.3 841 804.9 956.8 567.1 580.2 657.2 517.6 559 569.4 620.3 650.5 782.9 556.5 734.1 660.4 661.3 Cl/10−6 6037.2 6000 3860.6 4979.2 4420.3 5220.8 4230.2 5057 5537.6 3815.5 4061.6 4257.9 5084 5536.1 6376.4 4519.6 4159.3 6186.9 5395 Ti/10−6 4742.1 4697.5 4834.1 4714.8 4530.7 4377.6 4777.1 4603 4734.4 4868.5 4826 4845.9 4641.3 4639.4 4567 4743.7 4663.3 4562.8 4811.9 V/10−6 131.8 110 128.4 124.9 122.7 100.9 122.7 129.6 128.4 118.9 119.6 117.4 139.2 127.3 117 119.8 131.9 129.2 125 Cr/10−6 99.5 84.6 94.9 88.2 88.8 76.1 90.1 100.1 95.2 91.6 90.8 90 97.6 92.6 85.8 88.6 94.6 89.8 93.4 Mn/10−6 478.4 505.3 527.1 463 496.2 404.1 507 475 493.8 513.2 510.5 542.4 489.8 503.1 492.1 509.3 458.6 623.8 529.6 Co/10−6 16.5 16.6 18.6 16 21.8 13.9 16.6 16.7 16.1 15.6 15.6 17.7 17.7 17.5 16 17 16.7 16 16.8 Ni/10−6 40.3 35.8 40.8 37.2 36.9 30.8 37.5 39.9 38.5 38.5 37.3 37.5 39.4 39.5 35.9 36.9 38.8 37.3 39 Cu/10−6 48.9 28.2 38.5 36.5 35 18.9 26.3 40.1 28.6 28.9 25.4 26.5 57.9 28.5 25.2 26 44.3 43.9 27 Zn/10−6 130.8 95.9 117.6 113.9 110.4 83.4 103.3 126.8 107.2 108.3 103.2 105.5 154.7 107.4 95.7 102.5 135.4 129.5 105.4 Ga/10−6 21.2 19.7 22.5 20.2 20.9 18.3 21 22.9 21.6 22.8 20.9 21.6 21.3 21.7 19 21.5 22.2 20.7 21.7 As/10−6 17 15.5 19.4 12.3 10.5 8.3 12.2 14.8 12.5 12.6 12.2 13.2 17.3 11.6 9.2 11.5 14 14.7 11.7 Rb/10−6 150.6 135.9 153.3 141.8 140.8 122.5 152.7 163.4 158 157.3 151.8 152.6 150.5 151.1 137.3 150.6 149.7 144.6 154 Sr/10−6 144 149 149 149 170 149 149 155 159 151 152 147 151 162 167 152 151 155 155 Zr/10−6 194 208 190 203 189 236 196 170 177 192 198 196 184 179 200 194 189 191 187 Nb/10−6 18.3 21.6 18 17.3 17.7 17 18.8 17.7 20.4 18.5 19 18.7 17.8 19 18.9 18.5 21.1 17.8 18.2 U/10−6 3.7 3.8 3.5 3.5 3.9 2.9 3.8 3.9 4.2 4.1 3.8 3.8 3.8 3.8 3.4 3.8 3.9 3.6 3.6 Mo/10−6 1.2 1.5 1.3 1.1 1.3 1 1.4 1.5 1.6 1.3 1.3 1.3 1.4 1.4 1.3 1.3 1.7 1.2 1.3 Sn/10−6 10.1 9.7 8.3 7.3 10.6 8 12.2 14.6 13.6 9.5 11.1 14.2 10.9 14.4 15.2 10.8 18.7 13.6 10.9 Sb/10−6 15.2 9.6 9.3 7.1 10 11.2 13.4 18.3 13.8 7.7 12.3 15.7 11.8 20.6 17.2 13 24.5 15 14.9 Ba/10−6 524 474 521 493 507 430 517 549 515 509 500 497 562 504 487 503 537 518 500 Hf/10−6 5.5 5.9 5.4 6 5.5 6.7 5.5 5.1 5.1 5.7 5.9 5.5 5.3 5.4 5.8 5.5 5.2 5.6 5.3 W/10−6 2.9 7.3 5.1 12.1 9.1 12.9 4.4 2.4 6.3 4.3 4.2 6.9 11.6 3.2 7.6 4.3 4.4 6.6 4.6 Pb/10−6 93.5 33.6 55.9 58.3 49.9 27.6 36.8 61.1 37.5 37.4 33.8 34.5 116.9 39.3 36.7 38.7 92.3 94.9 36 Bi/10−6 0.5 0.2 1 1.3 1.4 2.4 1.2 1.6 0.3 1.3 1.5 0 0 0 0.7 1 0.6 1.3 0 Th/10−6 15.2 15 13.8 13.9 12.8 10.6 16.1 14 16.1 14.7 14.6 15.5 14.7 16.4 13.1 14.8 15.1 14.7 14.5 Ce/10−6 76.5 54.2 62.8 73.7 71.8 74.8 65.5 68.4 78.7 72.8 66.4 69.4 66.8 68.4 72.9 65.1 68 65.7 84.4 Nd/10−6 27.8 38.4 34 29.1 32.8 29.3 35.1 31 26.9 26.8 33.6 30.8 35.7 31.3 31.8 28 27.5 25.2 29.5 Y/10−6 26.8 27.4 26.8 27 25.5 26.1 26.8 24.4 26 26.2 27.7 27.2 25.8 25.8 26.1 26.7 25.4 25.6 27 La/10−6 44.7 36.3 44.3 35.3 36.4 40.3 41.2 36.4 44.6 44.8 39.8 42 37.7 38.1 33.8 40.8 38.2 44.6 43.8 Sc/10−6 13.9 15 16.6 14.7 15.5 10.6 16 15.6 16.1 13.6 12.9 17.5 13.4 14.3 12.8 15.9 15.1 15.4 14.3 TN/% 0.16 0.1 0.14 0.1 0.13 0.07 0.11 0.15 0.13 0.12 0.1 0.15 0.16 0.12 0.13 0.12 0.14 0.14 0.12 TC/% 1.43 1.1 1.29 1.24 1.58 1.16 1.33 1.54 1.33 1.39 1.38 1.63 1.43 1.62 1.64 1.41 1.52 1.43 1.52 TOC/% 1.05 0.7 0.91 0.69 0.92 0.67 0.82 0.93 0.85 0.66 0.86 1.01 0.96 0.98 0.86 0.85 1.04 1.02 0.9 表 2 基于16S rRNA基因序列的菌株分类鉴定信息
Table 2. Isolation and identification of strains based on 16S rRNA gene sequence analysis
菌株号 亲缘菌株 16S rRNA
基因相似性/%层位深度
/cmbsf16S rRNA基因
序列登录号18A01 Alcanivorax xenomutans JC109T 99.22 18 OQ186762 18E01 Bacillus tequilensis KCTC 13622T 99.79 18 OQ186763 18E02 Pelagibacterium halotolerans B2T 99.78 18 OQ186764 18E03 Bacillus altitudinis 41KF2bT 99.93 18 OQ186765 18E04 Pseudonocardia carboxydivorans Y8T 99.93 18 OQ186766 18E05 Alcanivorax xenomutans JC109T 100 18 OQ186767 18M01 Alcanivorax venustensis ISO4T 100 18 OQ186768 18S01 Alcanivorax xenomutans JC109T 100 18 OQ186769 81A02 Alcanivorax xenomutans JC109T 99.93 81 OQ186770 81E01 Rossellomorea aquimaris TF-12T 99.21 81 OQ186771 81E02 Sutcliffiella halmapala DSM 8723T 99.14 81 OQ186772 81E03 Rossellomorea aquimaris TF-12T 99.29 81 OQ186773 81E05 Fictibacillus arsenicus Con a/3T 99.71 81 OQ186774 81E06 Bacillus safensis subsp. safensis FO-36bT 99.86 81 OQ186775 81E07-1 Alcanivorax xenomutans JC109T 99.93 81 OQ186777 81E07-2 Staphylococcus pseudoxylosus S04009T 99.86 81 OQ186778 81E08 Mesobacillus thioparans BMP-1T 99.71 81 OQ186779 81E09 Mesorhizobium sediminum YIM M12096T 99.77 81 OQ186780 110E01 Halomonas titanicae BH1T 99.44 110 OQ186781 110E02 Metabacillus idriensis SMC 4352-2T 99.93 110 OQ186782 110E03 Pseudonocardia carboxydivorans Y8T 100 110 OQ186783 110E04 Mesorhizobium sediminum YIM M12096T 99.77 110 OQ186784 110E05 Pelagibacterium halotolerans B2T 99.78 110 OQ186785 110E06 Sutcliffiella horikoshii DSM 8719T 99.29 110 OQ186786 110E07 Virgibacillus halodenitrificans DSM 10037T 99.93 110 OQ186787 181E01 Halomonas titanicae BH1T 100 181 OQ186788 181E02 Rossellomorea aquimaris TF-12T 99.36 181 OQ186789 181E03 Pseudonocardia carboxydivorans Y8T 99.79 181 OQ186790 181E04 Nitratireductor aquibiodomus JCM 21793T 99.08 181 OQ186791 181M01 Halomonas titanicae BH1T 100 181 OQ186792 181S01 Martelella mediterranea DSM 17316T 99.63 181 OQ186793 199A01 Alcanivorax xenomutans JC109T 100 199 OQ186794 199E01 Nitratireductor aquibiodomus JCM 21793T 99.17 199 OQ186795 199E02 Mesorhizobium sediminum YIM M12096T 99.77 199 OQ186796 199E03 Pseudonocardia carboxydivorans Y8T 99.78 199 OQ186797 199M01 Halomonas titanicae BH1T 100 199 OQ186798 248E02 Paenisporosarcina quisquiliarum SK 55T 99.38 248 OQ186799 248E04 Nitratireductor aquibiodomus JCM 21793T 99.03 248 OQ186800 248E05 Citromicrobium bathyomarinum JF-1T 99.7 248 OQ186801 301A01 Alcanivorax xenomutans JC109T 99.93 301 OQ186802 301E02 Paenisporosarcina macmurdoensis CMS 21wT 99.31 301 OQ186803 301E04 Paenisporosarcina macmurdoensis CMS 21wT 99.72 301 OQ186804 301E05 Pelagerythrobacter marinus H32T 99.93 301 OQ186805 301E06 Nitratireductor aquibiodomus JCM 21793T 98.94 301 OQ186806 334E03 Paenisporosarcina macmurdoensis CMS 21wT 99.15 334 OQ186807 334E04 Paenisporosarcina macmurdoensis CMS 21wT 99.14 334 OQ186808 334E05 Alcanivorax xenomutans JC109T 100 334 OQ186809 334M01 Alcanivorax xenomutans JC109T 99.93 334 OQ186810 334S02 Alcanivorax xenomutans JC109T 100 334 OQ186811 334S03 Muricauda ruestringensis DSM 13258T 98.48 334 OQ186812 334S04 Alcanivorax xenomutans JC109T 100 334 OQ186813 392A01 Alcanivorax xenomutans JC109T 99.93 392 OQ186814 392E01 Paenisporosarcina quisquiliarum SK 55T 99.71 392 OQ186815 392E02 Alcanivorax xenomutans JC109T 100 392 OQ186816 392E03 Pelagerythrobacter marinus H32T 100 392 OQ186817 392E04 Pelagibacterium nitratireducens JLT2005T 99.77 392 OQ186818 392E05 Paracoccus marcusii DSM 11574T 99.77 392 OQ186819 392E06 Paenisporosarcina quisquiliarum SK 55T 99.79 392 OQ186820 475E01 Alkalihalobacillus hwajinpoensis SW-72T 99.65 475 OQ186821 475E02 Thalassospira xiamenensis M-5T 99.65 475 OQ186822 475E03 Qipengyuania vulgaris 022 2-10T 99.85 475 OQ186823 475E04 Pelagibacterium nitratireducens JLT2005T 99.7 475 OQ186824 475S01 Alcanivorax xenomutans JC109T 99.93 475 OQ186825 475M01 Thalassospira xiamenensis M-5T 99.85 475 OQ186826 -
[1] 王风平, 周悦恒, 张新旭, 等. 深海微生物多样性[J]. 生物多样性, 2013, 21(4): 446-456
WANG Fengping, ZHOU Yueheng, ZHANG Xinxu, et al. Biodiversity of deep-sea microorganisms[J]. Biodiversity Science, 2013, 21(4): 446-456.
[2] 李学恭, 徐俊, 肖湘. 深海微生物高压适应与生物地球化学循环[J]. 微生物学通报, 2013, 40(1): 59-70
LI Xuegong, XU Jun, XIAO Xiang. High pressure adaptation of deep-sea microorganisms and biogeochemical cycles[J]. Microbiology China, 2013, 40(1): 59-70.
[3] Takai K, Nakagawa S, Nunoura T. Comparative investigation of microbial communities associated with hydrothermal activities in the Okinawa Trough[M]//Ishibashi J I, Okino K, Sunamura M. Subseafloor Biosphere Linked to Hydrothermal Systems. Tokyo: Springer, 2015: 421-435.
[4] Takai K, Nakamura K. Compositional, physiological and metabolic variability in microbial communities associated with geochemically diverse, deep-sea hydrothermal vent fluids[M]//Barton L L, Mandl M, Loy A. Geomicrobiology: Molecular and Environmental Perspective. Dordrecht: Springer, 2010: 251-283.
[5] Nakamura K, Takai K. Theoretical constraints of physical and chemical properties of hydrothermal fluids on variations in chemolithotrophic microbial communities in seafloor hydrothermal systems[J]. Progress in Earth and Planetary Science, 2014, 1(1): 5. doi: 10.1186/2197-4284-1-5
[6] Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance[J]. FEMS Microbiology Ecology, 2008, 65(1): 1-14. doi: 10.1111/j.1574-6941.2008.00502.x
[7] Takai K, Nakamura K. Archaeal diversity and community development in deep-sea hydrothermal vents[J]. Current Opinion in Microbiology, 2011, 14(3): 282-291. doi: 10.1016/j.mib.2011.04.013
[8] Guan L, Cho K H, Lee J H. Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria[J]. Food Microbiology, 2011, 28(1): 101-113. doi: 10.1016/j.fm.2010.09.001
[9] Thornburg C C, Zabriskie T M, McPhail K L. Deep-sea hydrothermal vents: potential hot spots for natural products discovery?[J]. Journal of Natural Products, 2010, 73(3): 489-499. doi: 10.1021/np900662k
[10] Wang S S, Jiang L J, Hu Q T, et al. Elemental sulfur reduction by a deep-sea hydrothermal vent Campylobacterium Sulfurimonas sp. NW10[J]. Environmental Microbiology, 2021, 23(2): 965-979. doi: 10.1111/1462-2920.15247
[11] 杜瑞, 于敏, 程景广, 等. 冲绳海槽热液区可培养硫氧化细菌多样性及其硫氧化特性[J]. 微生物学报, 2019, 59(6): 1036-1049 doi: 10.13343/j.cnki.wsxb.20180353
DU Rui, YU Min, CHENG Jingguang, et al. Diversity and sulfur oxidation characteristics of cultivable sulfur oxidizing bacteria in hydrothermal fields of Okinawa Trough[J]. Acta microbiologica Sinica, 2019, 59(6): 1036-1049. doi: 10.13343/j.cnki.wsxb.20180353
[12] Venter J C, Remington K, Heidelberg J F, et al. Environmental genome shotgun sequencing of the Sargasso Sea[J]. Science, 2004, 304(5667): 66-74. doi: 10.1126/science.1093857
[13] 刘明华, 王健鑫, 俞凯成, 等. 东海陆架表层沉积物细菌群落结构及地理分布研究[J]. 海洋与湖沼, 2015, 46(5): 1119-1131
LIU Minghua, WANG Jianxin, YU Kaicheng, et al. Community structure and geographical distribution of bacterial on surface layer sediments in the east China Sea[J]. Oceanologia et Limnologia Sinica, 2015, 46(5): 1119-1131.
[14] 王苑如, 崔鸿鹏, 李继东, 等. 西太平洋多金属结核区表层沉积物细菌群落结构及其对沉积扰动的响应[J]. 海洋学研究, 2021, 39(2): 21-32
WANG Yuanru, CUI Hongpeng, LI Jidong, et al. The structure of bacterial communities and its response to the sedimentary disturbance in the surface sediment of western Pacific polymetallic nodule area[J]. Journal of Marine Sciences, 2021, 39(2): 21-32.
[15] Inagaki F, Kuypers M M M, Tsunogai U, et al. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(38): 14164-14169.
[16] Yanagawa K, Ijiri A, Breuker A, et al. Defining boundaries for the distribution of microbial communities beneath the sediment-buried, hydrothermally active seafloor[J]. The ISME Journal, 2017, 11(2): 529-542. doi: 10.1038/ismej.2016.119
[17] Nakagawa S, Takai K, Inagaki F, et al. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation[J]. FEMS Microbiology Ecology, 2005, 54(1): 141-155. doi: 10.1016/j.femsec.2005.03.007
[18] Wang L, Yu M, Liu Y, et al. Comparative analyses of the bacterial community of hydrothermal deposits and seafloor sediments across Okinawa Trough[J]. Journal of Marine Systems, 2018, 180: 162-172. doi: 10.1016/j.jmarsys.2016.11.012
[19] Yanagawa K, Breuker A, Schippers A, et al. Microbial community stratification controlled by the subseafloor fluid flow and geothermal gradient at the Iheya North hydrothermal field in the Mid-Okinawa Trough (Integrated Ocean Drilling Program Expedition 331)[J]. Applied and Environmental Microbiology, 2014, 80(19): 6126-6135. doi: 10.1128/AEM.01741-14
[20] Yanagawa K, Morono Y, de Beer D, et al. Metabolically active microbial communities in marine sediment under high-CO2 and low-pH extremes[J]. The ISME Journal, 2013, 7(3): 555-567. doi: 10.1038/ismej.2012.124
[21] Zhang J, Sun Q L, Zeng Z G, et al. Microbial diversity in the deep-sea sediments of Iheya North and Iheya Ridge, Okinawa Trough[J]. Microbiological Research, 2015, 177: 43-52. doi: 10.1016/j.micres.2015.05.006
[22] Rona P A, Scott S D. A special issue on sea-floor hydrothermal mineralization: new perspectives: preface[J]. Economic Geology, 1993, 88(8): 1935-1976. doi: 10.2113/gsecongeo.88.8.1935
[23] 邵珂, 陈建平, 任梦依. 印度洋中脊多金属硫化物矿产资源定量预测与评价[J]. 海洋地质与第四纪地质, 2015, 35(5): 125-133
SHAO Ke, CHEN Jianping, REN Mengyi. Quantitative prediction and evaluation of polymetallic sulfide mineral deposits along the central Indian ocean ridge[J]. Marine Geology & Quaternary Geology, 2015, 35(5): 125-133.
[24] Walsh J J. Importance of continental margins in the marine biogeochemical cycling of carbon and nitrogen[J]. Nature, 1991, 350(6313): 53-55. doi: 10.1038/350053a0
[25] 胡思谊. 冲绳海槽南部S3岩心沉积物的矿物学和地球化学研究[D]. 中国科学院大学(中国科学院海洋研究所)博士学位论文, 2020
HU Siyi. Mineralogical and geochemical study of sediment core S3 from the southern Okinawa Trough[D]. Doctor Dissertation of Institute of Oceanology, Chinese Academy of Sciences, 2020.
[26] 杨宝菊, 吴永华, 刘季花, 等. 冲绳海槽表层沉积物元素地球化学及其对物源和热液活动的指示[J]. 海洋地质与第四纪地质, 2018, 38(2): 25-37
YANG Baoju, WU Yonghua, LIU Jihua, et al. Elemental geochemistry of surface sediments in Okinawa Trough and its implications for provenance and hydrothermal activity[J]. Marine Geology & Quaternary Geology, 2018, 38(2): 25-37.
[27] 赵维殳, 肖湘. 深海热液区微生物群落与环境适应性机理[J]. 中国科学: 生命科学, 2023, 53(5): 660-671
ZHAO Weishu, XIAO Xiang. Microbiome and environmental adaption mechanisms in deep-sea hydrothermal vents[J]. Scientia Sinica: Vitae, 2023, 53(5): 660-671.
[28] 杨娅敏. 冲绳海槽南部浊流沉积层中的硫化物特征研究[D]. 中国科学院大学(中国科学院海洋研究所)博士学位论文, 2021
YANG Yamin. Study on the characteristics of turbidite sediments hosted-sulfides deposit from the southern Okinawa Trough[D]. Doctor Dissertation of Institute of Oceanology, Chinese Academy of Sciences, 2021.
[29] Zeng Z G, Chen S, Ma Y, et al. Chemical compositions of mussels and clams from the Tangyin and Yonaguni Knoll IV hydrothermal fields in the southwestern Okinawa Trough[J]. Ore Geology Reviews, 2017, 87: 172-191. doi: 10.1016/j.oregeorev.2016.09.015
[30] 尚鲁宁, 陈磊, 张训华, 等. 冲绳海槽南部海底热液活动区地形地貌特征及成因分析[J]. 海洋地质与第四纪地质, 2019, 39(4): 12-22
SHANG Luning, CHEN Lei, ZHANG Xunhua, et al. Topographic features of the hydrothermal field and their genetic mechanisms in southern Okinawa Trough[J]. Marine Geology & Quaternary Geology, 2019, 39(4): 12-22.
[31] Atlas R M. Handbook of Microbiological Media[M]. Boca Raton: CRC Press, 1993: 527.
[32] Ruby E G, Wirsen C O, Jannasch H W. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos rift hydrothermal vents[J]. Applied and Environmental Microbiology, 1981, 42(2): 317-324. doi: 10.1128/aem.42.2.317-324.1981
[33] Das A, Cao W R, Zhang H J, et al. Carbon fixation in sediments of Sino-Pacific seas-differential contributions of bacterial and archaeal domains[J]. Journal of Marine Systems, 2017, 175: 15-23. doi: 10.1016/j.jmarsys.2017.06.005
[34] Cao W R, Lu D C, Sun X K, et al. Seonamhaeicola maritimus sp. nov., isolated from coastal sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(2): 902-908.
[35] Wang F, Men X, Zhang G, et al. Assessment of 16S rRNA gene primers for studying bacterial community structure and function of aging flue-cured tobaccos[J]. AMB Express, 2018, 8(1): 182. doi: 10.1186/s13568-018-0713-1
[36] Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Molecular Biology and Evolution, 2021, 38(7): 3022-3027. doi: 10.1093/molbev/msab120
[37] Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach[J]. Journal of Molecular Evolution, 1981, 17(6): 368-376. doi: 10.1007/BF01734359
[38] Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences[J]. Journal of Molecular Evolution, 1980, 16(2): 111-120. doi: 10.1007/BF01731581
[39] Nossa C W, Oberdorf W E, Yang L Y, et al. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome[J]. World Journal of Gastroenterology, 2010, 16(33): 4135-444. doi: 10.3748/wjg.v16.i33.4135
[40] Yang Y M, Zeng Z G, Yin X B, et al. Mineralogy, geochemistry, and sulfur isotope characteristics of sediment-hosted hydrothermal sulfide minerals from the southern Okinawa Trough[J]. Acta Oceanologica Sinica, 2021, 40(10): 129-143. doi: 10.1007/s13131-021-1836-9
[41] 赵一阳, 翟世奎, 李永植, 等. 冲绳海槽中部热水活动的新记录[J]. 科学通报, 1996, 41(14): 1307-1310
ZHAO Yiyang, ZHAI Shikui, LI Yongzhi, et al. New records of submarine hydrothermal activity in middle part of the Okinawa Trough[J]. Chinese Science Bulletin, 1997, 42(7): 574-577.
[42] 蒋富清, 李安春, 李铁刚. 冲绳海槽北端表层沉积物过渡元素地球化学特征[J]. 海洋与湖沼, 2006, 37(1): 75-83 doi: 10.3321/j.issn:0029-814X.2006.01.012
JIANG Fuqing, LI Anchun, LI Tiegang. Geochemical characteristics of transition elements in surface sediments northern Okinawa Trough[J]. Oceanologia et Limnologia Sinica, 2006, 37(1): 75-83. doi: 10.3321/j.issn:0029-814X.2006.01.012
[43] Edlund A, Hårdeman F, Jansson J K, et al. Active bacterial community structure along vertical redox gradients in Baltic Sea sediment[J]. Environmental Microbiology, 2008, 10(8): 2051-2063. doi: 10.1111/j.1462-2920.2008.01624.x
[44] Chu H Y, Sun H B, Tripathi B M, et al. Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Xizang Plateau[J]. Environmental Microbiology, 2016, 18(5): 1523-1533. doi: 10.1111/1462-2920.13236
[45] Teske A P. Microbial community composition in deep marine subsurface sediments of ODP Leg 201: sequencing surveys and cultivations[M]//Jørgensen B B, D'Hondt S L, Miller D J. Proceedings of the Ocean Drilling Program Scientific Results. 2006.
[46] Pérez-Rodríguez I, Bohnert K A, Cuebas M, et al. Detection and phylogenetic analysis of the membrane-bound nitrate reductase (Nar) in pure cultures and microbial communities from deep-sea hydrothermal vents[J]. FEMS Microbiology Ecology, 2013, 86(2): 256-267. doi: 10.1111/1574-6941.12158
[47] Rajasabapathy R, Mohandass C, Colaco A, et al. Culturable bacterial phylogeny from a shallow water hydrothermal vent of Espalamaca (Faial, Azores) reveals a variety of novel taxa[J]. Current Science, 2014, 106(1): 58-69.
[48] Martins A, Tenreiro T, Andrade G, et al. Photoprotective bioactivity present in a unique marine bacteria collection from Portuguese deep sea hydrothermal vents[J]. Marine Drugs, 2013, 11(5): 1506-1523. doi: 10.3390/md11051506
[49] Kaye J Z, Sylvan J B, Edwards K J, et al. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments[J]. FEMS Microbiology Ecology, 2011, 75(1): 123-133. doi: 10.1111/j.1574-6941.2010.00984.x
[50] Cao H L, Wang Y, Lee O O, et al. Microbial sulfur cycle in two hydrothermal chimneys on the southwest Indian Ridge[J]. mBio, 2014, 5(1): e00980-13.
[51] Mohandass C, Rajasabapathy R, Ravindran C, et al. Bacterial diversity and their adaptations in the shallow water hydrothermal vent at D. João de Castro Seamount (DJCS), Azores, Portugal[J]. Cahiers de Biologie Marine, 2012, 53(1): 65-76.
[52] Dick G J, Lee Y E, Tebo B M. Manganese(II)-oxidizing Bacillus spores in Guaymas basin hydrothermal sediments and plumes[J]. Applied and Environmental Microbiology, 2006, 72(5): 3184-3190. doi: 10.1128/AEM.72.5.3184-3190.2006
[53] Liao L, Xu X W, Jiang X W, et al. Microbial diversity in deep-sea sediment from the cobalt-rich crust deposit region in the Pacific Ocean[J]. FEMS Microbiology Ecology, 2011, 78(3): 565-585. doi: 10.1111/j.1574-6941.2011.01186.x
[54] Xu M X, Wang F P, Meng J, et al. Construction and preliminary analysis of a metagenomic library from a deep-sea sediment of East Pacific Nodule province[J]. FEMS Microbiology Ecology, 2007, 62(3): 233-241. doi: 10.1111/j.1574-6941.2007.00377.x
[55] 魏曼曼. 劳盆地深海热液喷口沉积物中微生物群落结构研究[D]. 中南大学硕士学位论文, 2010
WEI Manman. Study on microbial community structures in sediments from Lau Basin deep-sea hydrothermal vents[D]. Master Dissertation of Central South University, 2010.
[56] Flores G E, Campbell J H, Kirshtein J D, et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge[J]. Environmental Microbiology, 2011, 13(8): 2158-2171. doi: 10.1111/j.1462-2920.2011.02463.x
[57] Sylvan J B, Toner B M, Edwards K J. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides[J]. mBio, 2012, 3(1): e00279-11.
[58] Cao W R, Wang L S, Saren G, et al. Variable microbial communities in the non-hydrothermal sediments of the Mid-Okinawa Trough[J]. Geomicrobiology Journal, 2020, 37(8): 774-782. doi: 10.1080/01490451.2020.1776800
[59] 张伟. 热带西太平洋深海沉积物微生物多样性及群落结构特征研究[D]. 中国科学院研究生院(海洋研究所)硕士学位论文, 2010
ZHANG Wei. Microbial diversity and community structure in marine sediments from the tropical western Pacific[D]. Master Dissertation of Institute of Oceanology, Chinese Academy of Sciences, 2010.
[60] Lee S Y, Huh C A, Su C C, et al. Sedimentation in the Southern Okinawa Trough: enhanced particle scavenging and teleconnection between the Equatorial Pacific and western Pacific margins[J]. Deep Sea Research Part I: Oceanographic Research Papers, 2004, 51(11): 1769-1780. doi: 10.1016/j.dsr.2004.07.008
[61] 邵宗泽. 深海热液区化能自养微生物多样性、代谢特征与环境作用[J]. 科学通报, 2018, 63(36): 3902-3910 doi: 10.1360/N972018-00980
SHAO Zongze. Diversity and metabolism of prokaryotic chemoautotrophs and their interactions with deep-sea hydrothermal environments[J]. Chinese Science Bulletin, 2018, 63(36): 3902-3910. doi: 10.1360/N972018-00980
[62] Li Y X, Li F C, Zhang X W, et al. Vertical distribution of bacterial and archaeal communities along discrete layers of a deep-sea cold sediment sample at the East Pacific Rise (~13°N)[J]. Extremophiles, 2008, 12(4): 573-585. doi: 10.1007/s00792-008-0159-5
[63] Wang P, Xiao X, Wang F P. Phylogenetic analysis of Archaea in the deep-sea sediments of west Pacific Warm Pool[J]. Extremophiles, 2005, 9(3): 209-217. doi: 10.1007/s00792-005-0436-5
-


 下载:
下载: