Advances in the study of methane-metabolizing microbial communities in marine sediments
-
摘要:
甲烷是一种重要的温室气体,深刻影响着全球的气候变化。同时,甲烷还是海底潜在能源—天然气水合物的主要成分。海洋沉积物是甲烷生物转化的一个重要生态区域,产甲烷菌主要利用H2、CO2及简单的有机物(甲醇、甲胺、二甲基硫等)作为底物生成甲烷,产生的甲烷在向上迁移的过程中主要被甲烷厌氧氧化(anaerobic oxidation of methane, AOM)和甲烷好氧氧化(aerobic oxidation of methane,AeOM)消耗,进而大大减少了甲烷向大气的排放量。AeOM主要发生在含氧的沉积物及沉积物-水界面中,由甲烷好氧氧化菌(aerobic methane-oxidizing bacteria, MOB)介导。然而,绝大部分甲烷在穿透缺氧沉积物层之前是被AOM反应消耗,甲烷厌氧氧化古菌(anaerobic methanotrophic archaea,ANME)是主要的参与者,这些功能微生物耦联电子受体SO42−、NO2−/NO3−或Fe3+和Mn4+将甲烷进行氧化。本文对产甲烷菌和甲烷氧化菌的种类、代谢途径及其在海洋沉积物中的分布特征进行了综述,并在前人工作基础上,对今后海洋生境中甲烷代谢过程的研究进行了展望,以期为进一步开展海洋环境中甲烷的生物转化过程及元素耦合的研究提供理论依据。
Abstract:Methane is an important greenhouse gas affecting the global climate. Meanwhile, methane is a major component of natural gas hydrate which regarded as a potential energy resource below seafloor. Seafloor sediment is an important ecological region for methane biotransformation. The methanogens can use H2, CO2, and simple organic compounds (e.g. methanol, methylamines, dimethylsulfide) as substrates to produce methane. The methane produced in the bottom of the sediments would be consumed by aerobic methanotrophs and anaerobic methanotrophs during its upward migration, which reduces greatly the methane emissions to the atmosphere. Aerobic methane oxidation occurs mainly in oxygenated sediments and sediment-water interfaces, and is mediated by aerobic methane-oxidizing bacteria. However, most of the methane is consumed by anaerobic methane oxidation before it reaches the seafloor. The anaerobic methanotrophs oxidize methane coupled by SO42−, NO2−/NO3− or Fe3+/Mn4+. We reviewed the status quo and perspectives of the taxonomy, metabolic and ecological diversity of methanogens and methanotrophs in marine sediments, and emphasized deficiencies and issues need to be solved in future studies. This review provided theoretical foundation for the study of biotransformation process and element coupling of methane in marine environment.
-
Key words:
- methane /
- marine sediment /
- methanogen /
- methanotrophs
-
-
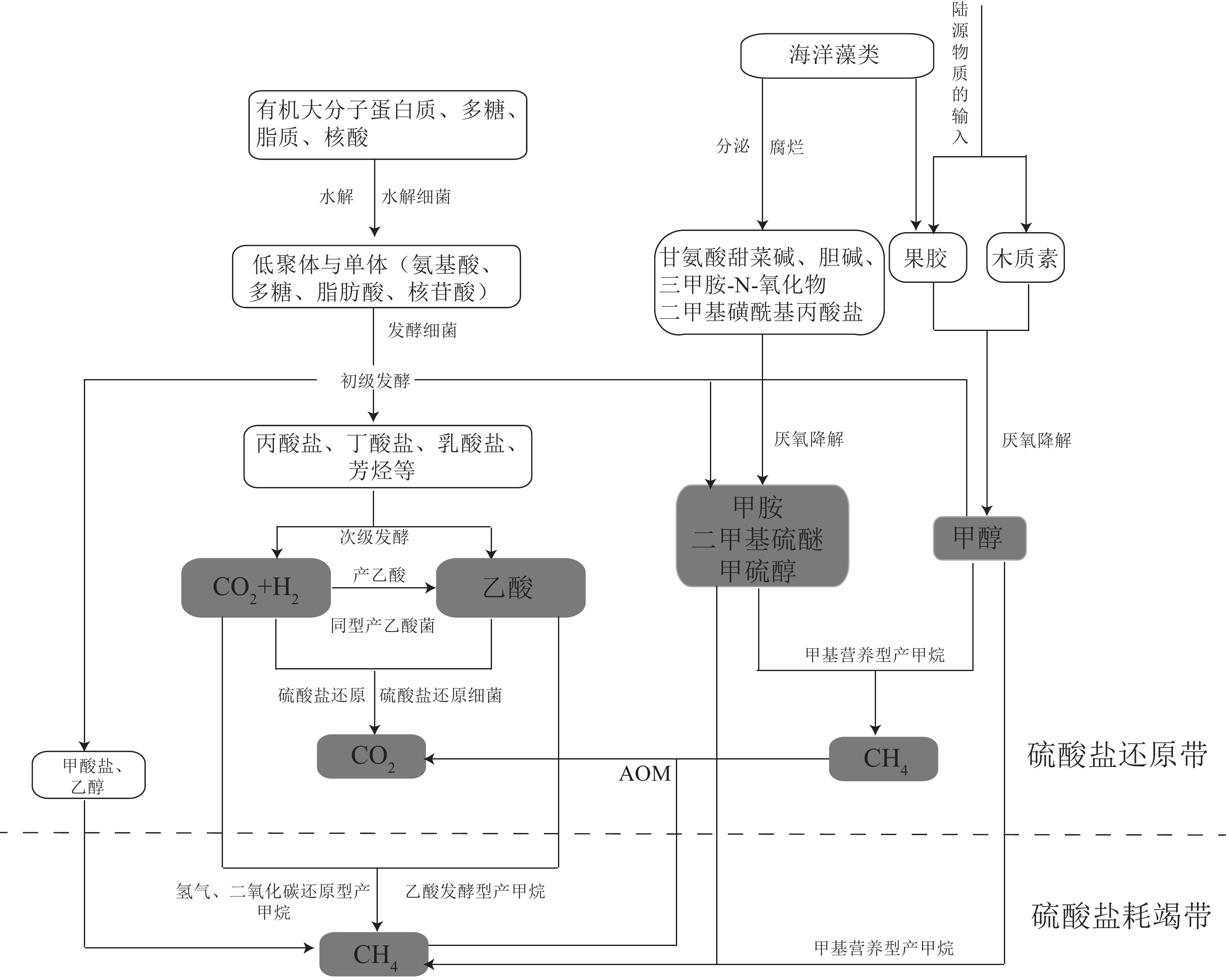
图 2 海洋沉积物中低分子化合物的生成和产甲烷途径 [22]
Figure 2.
图 4 基于mcrA序列的ANME古菌系统发育树 [40]
Figure 4.
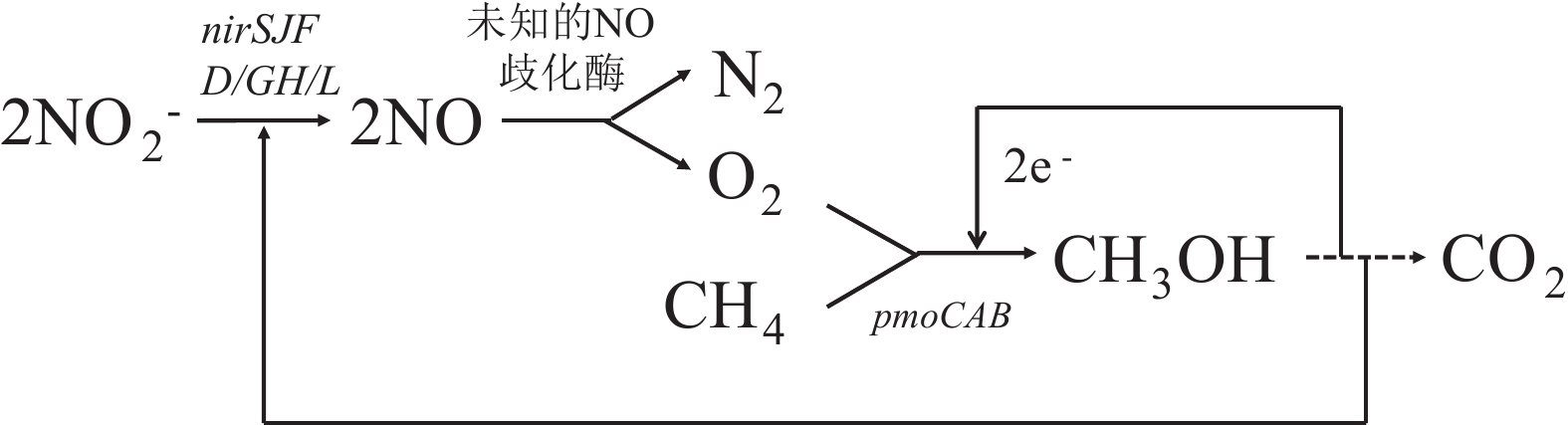
图 5 Methylomirabilis oxyfera 的DAMO理论途径[46]
Figure 5.
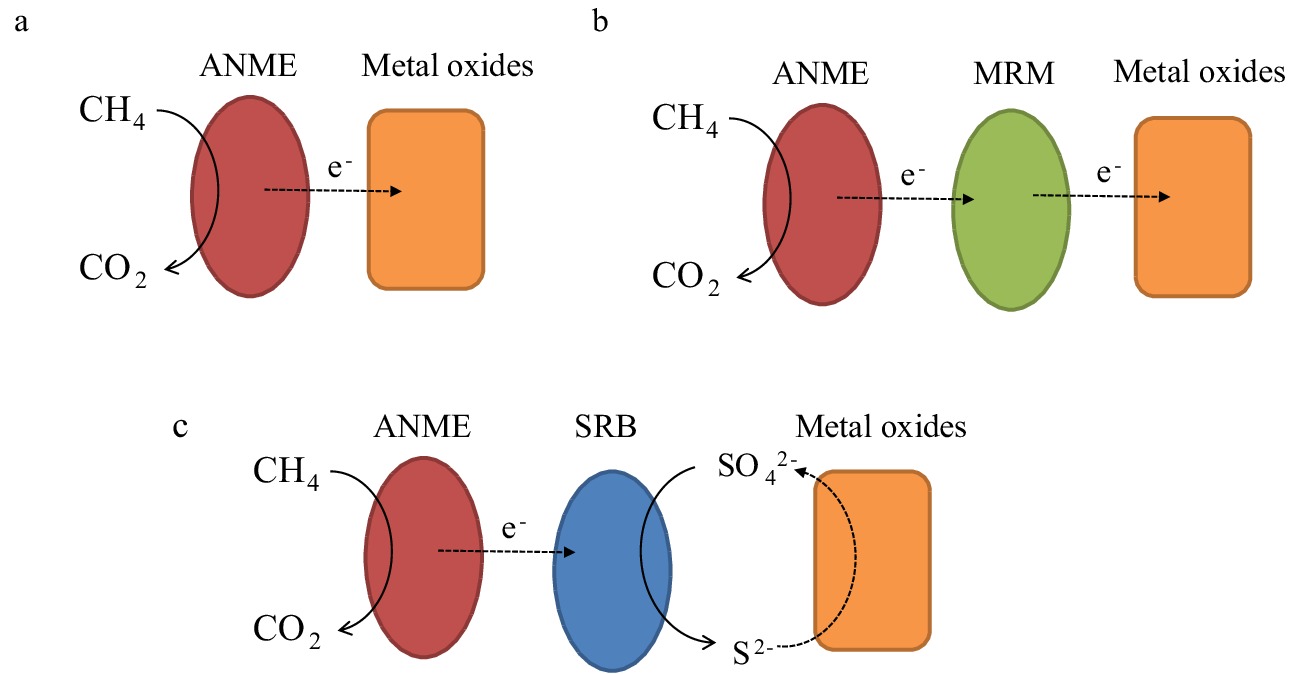
图 6 微生物介导Metal-AOM的不同反应机制[76]
Figure 6.
表 1 不同电子受体类型甲烷氧化反应的吉布斯自由能[45-46]
Table 1. Standard Gibbs free energies with different electron acceptors for methane oxidation [45-46]
不同电子受体介导的甲烷氧化反应 吉布斯自由能/(kJ·mol−1 CH4) CH4+2O2→CO2+2H2O −858.7 CH4+SO42−→HCO3−+HS−+H2O −33.0 CH4+4NO3−→HCO3−+4NO2−+H++H2O −483.4 3CH4+8NO2−+8H+→ 4N2+3CO2+10H2O −928.0 CH4+8Fe3++2H2O→CO2+8Fe2++8H+ −471.0 5CH4+8MnO4−+19H+→5HCO3-+8Mn2++17H2O −1 008.1 -
[1] IPCC. Climate Change 2014: Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change[M]. Cambridge, UK: Cambridge University Press, 2004.
[2] Rice D D. Biogenic gas: controls, habitats, and resource potential[M]//Howell D G. The Future of Energy Gases. Washington: United States Government Printing Office, 1993: 583-606.
[3] Hinrichs K U, Boetius A. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry[M]//Wefer G, Billett D, Hebbeln D, et al. Ocean Margin Systems. Berlin: Springer, 2002: 457-477.
[4] Reeburgh W S. Oceanic methane biogeochemistry [J]. Chemical Reviews, 2007, 107(2): 486-513. doi: 10.1021/cr050362v
[5] Reeburgh W S. Methane consumption in Cariaco Trench waters and sediments [J]. Earth and Planetary Science Letters, 1976, 28(3): 337-344. doi: 10.1016/0012-821X(76)90195-3
[6] Islas-Lima S, Thalasso F, Gómez-Hernandez J. Evidence of anoxic methane oxidation coupled to denitrification [J]. Water Research, 2004, 38(1): 13-16. doi: 10.1016/j.watres.2003.08.024
[7] Beal E J, House C H, Orphan V J. Manganese- and iron-dependent marine methane oxidation [J]. Science, 2009, 325(5937): 184-187. doi: 10.1126/science.1169984
[8] Kallmeyer J, Pockalny R, Adhikari R R, et al. Global distribution of microbial abundance and biomass in subseafloor sediment [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(40): 16213-16216. doi: 10.1073/pnas.1203849109
[9] Liu Y C, Whitman W B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea [J]. Annals of the New York Academy of Sciences, 2008, 1125(1): 171-189. doi: 10.1196/annals.1419.019
[10] Adam P S, Borrel G, Brochier-Armanet C, et al. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology [J]. The ISME Journal, 2017, 11(11): 2407-2425. doi: 10.1038/ismej.2017.122
[11] Bapteste É, Brochier C, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens [J]. Archaea, 2005, 2005: 859728.
[12] Borrel G, Parisot N, Harris H M, et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine [J]. BMC Genomics, 2014, 15: 679. doi: 10.1186/1471-2164-15-679
[13] Nobu M K, Narihiro T, Kuroda K, et al. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen [J]. The ISME Journal, 2016, 10(10): 2478-2487. doi: 10.1038/ismej.2016.33
[14] Sorokin D Y, Makarova K S, Abbas B, et al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis [J]. Nature Microbiology, 2017, 2(8): 17081. doi: 10.1038/nmicrobiol.2017.81
[15] Evans P N, Parks D H, Chadwick G L, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics [J]. Science, 2015, 350(6259): 434-438. doi: 10.1126/science.aac7745
[16] Vanwonterghem I, Evans P N, Parks D H, et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota [J]. Nature Microbiology, 2016, 1: 16170. doi: 10.1038/nmicrobiol.2016.170
[17] Wang Y L, Hua Z S, Goh K M, et al. Further expansion of methane metabolism in the Archaea. BioRxiv, 2018.
[18] Conklin A, Stensel H D, Ferguson J. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion [J]. Water Environment Research, 2006, 78(5): 486-496. doi: 10.2175/106143006X95393
[19] Kobayashi T, Yasuda D, Li Y Y, et al. Characterization of start-up performance and archaeal community shifts during anaerobic self-degradation of waste-activated sludge [J]. Bioresource Technology, 2009, 100(21): 4981-4988. doi: 10.1016/j.biortech.2009.05.043
[20] 段昌海, 张翠景, 孙艺华, 等. 新型产甲烷古菌研究进展[J]. 微生物学报, 2019, 59(6):981-995
DUAN Changhai, ZHANG Cuijing, SUN Yihua, et al. Recent advances on the novel methanogens [J]. Acta Microbiologica Sinica, 2019, 59(6): 981-995.
[21] Zhou Z, Zhang C J, Liu P F, et al. Non-syntrophic methanogenic hydrocarbon degradation by an archaeal species [J]. Nature, 2022, 601(7892): 257-262. doi: 10.1038/s41586-021-04235-2
[22] Konhauser K O. Introduction to Geomicrobiology[M]. John Wiley & Sons, 2009.
[23] Xiao K Q, Beulig F, Kjeldsen K U, et al. Concurrent methane production and oxidation in surface sediment from Aarhus Bay, Denmark [J]. Frontiers in Microbiology, 2017, 8: 1198. doi: 10.3389/fmicb.2017.01198
[24] Xiao K Q, Beulig F, Røy H, et al. Methylotrophic methanogenesis fuels cryptic methane cycling in marine surface sediment [J]. Limnology and Oceanography, 2018, 63(4): 1519-1527. doi: 10.1002/lno.10788
[25] Zhuang G C, Heuer V B, Lazar C S, et al. Relative importance of methylotrophic methanogenesis in sediments of the Western Mediterranean Sea [J]. Geochimica et Cosmochimica Acta, 2018, 224: 171-186. doi: 10.1016/j.gca.2017.12.024
[26] Oremland R S, Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments [J]. Applied and Environmental Microbiology, 1982, 44(6): 1270-1276. doi: 10.1128/aem.44.6.1270-1276.1982
[27] Li L Y, Zhang W T, Zhang S J, et al. Bacteria and archaea synergistically convert glycine betaine to biogenic methane in the Formosa cold seep of the South China sea [J]. Msystems, 2021, 6(5): e0070321. doi: 10.1128/mSystems.00703-21
[28] Dolfing J, Larter S R, Head I M. Thermodynamic constraints on methanogenic crude oil biodegradation [J]. The ISME Journal, 2008, 2(4): 442-452. doi: 10.1038/ismej.2007.111
[29] Ozuolmez D, Na H, Lever M A, et al. Methanogenic archaea and sulfate reducing bacteria co-cultured on acetate: teamwork or coexistence? [J]. Frontiers in Microbiology, 2015, 6: 492.
[30] Chen Y, Wu N Y, Liu C L, et al. Methanogenesis pathways of methanogens and their responses to substrates and temperature in sediments from the South Yellow Sea [J]. Science of the Total Environment, 2022, 815: 152645. doi: 10.1016/j.scitotenv.2021.152645
[31] Hanson R S, Hanson T E. Methanotrophic bacteria [J]. Microbiological Reviews, 1996, 60(2): 439-471. doi: 10.1128/mr.60.2.439-471.1996
[32] Dedysh S N, Knief C, Dunfield P F. Methylocella species are facultatively methanotrophic [J]. Journal of Bacteriology, 2005, 187(13): 4665-4670. doi: 10.1128/JB.187.13.4665-4670.2005
[33] Vorobev A V, Baani M, Doronina N V, et al. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase [J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(10): 2456-2463. doi: 10.1099/ijs.0.028118-0
[34] Elsaied H E, Hayashi T, Naganuma T. Molecular analysis of deep-sea hydrothermal vent aerobic methanotrophs by targeting genes of 16S rRNA and particulate methane monooxygenase [J]. Marine Biotechnology, 2004, 6(5): 503-509. doi: 10.1007/s10126-004-3042-0
[35] Tavormina P L, Ussler III W, Orphan V J. Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American margin [J]. Applied and Environmental Microbiology, 2008, 74(13): 3985-3995. doi: 10.1128/AEM.00069-08
[36] Wasmund K, Kurtböke D I, Burns K A, et al. Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity [J]. FEMS Microbiology Ecology, 2009, 68(2): 142-151. doi: 10.1111/j.1574-6941.2009.00667.x
[37] Alperin M J, Hoehler T M. Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates: 1. Thermodynamic and physical constraints [J]. American Journal of Science, 2009, 309(10): 869-957. doi: 10.2475/10.2009.01
[38] Knittel K, Lösekann T, Boetius A, et al. Diversity and distribution of methanotrophic archaea at cold seeps [J]. Applied and Environmental Microbiology, 2005, 71(1): 467-479. doi: 10.1128/AEM.71.1.467-479.2005
[39] Haroon M F, Hu S H, Shi Y, et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage [J]. Nature, 2013, 500(7464): 567-570. doi: 10.1038/nature12375
[40] Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process [J]. Annual Review of Microbiology, 2009, 63: 311-334. doi: 10.1146/annurev.micro.61.080706.093130
[41] Timmers P H A, Widjaja-Greefkes H C A, Plugge C M, et al. Evaluation and optimization of PCR primers for selective and quantitative detection of marine ANME subclusters involved in sulfate-dependent anaerobic methane oxidation [J]. Applied Microbiology and Biotechnology, 2017, 101(14): 5847-5859. doi: 10.1007/s00253-017-8338-x
[42] Kong Y, Lei H Y, Zhang Z L, et al. Depth profiles of geochemical features, geochemical activities and biodiversity of microbial communities in marine sediments from the Shenhu area, the northern South China Sea [J]. Science of the Total Environment, 2021, 779: 146233. doi: 10.1016/j.scitotenv.2021.146233
[43] Yang S S, Lv Y X, Liu X P, et al. Genomic and enzymatic evidence of acetogenesis by anaerobic methanotrophic archaea [J]. Nature Communications, 2020, 11(1): 3941. doi: 10.1038/s41467-020-17860-8
[44] Metcalfe K S, Murali R, Mullin S W, et al. Experimentally-validated correlation analysis reveals new anaerobic methane oxidation partnerships with consortium-level heterogeneity in diazotrophy [J]. The ISME Journal, 2021, 15(2): 377-396. doi: 10.1038/s41396-020-00757-1
[45] Caldwell S L, Laidler J R, Brewer E A, et al. Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms [J]. Environmental Science & Technology, 2008, 42(18): 6791-6799.
[46] Ettwig K F, Butler M K, Le Paslier D, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria [J]. Nature, 2010, 464(7288): 543-548. doi: 10.1038/nature08883
[47] Roalkvam I, Jørgensen S L, Chen Y F, et al. New insight into stratification of anaerobic methanotrophs in cold seep sediments [J]. FEMS Microbiology Ecology, 2011, 78(2): 233-243. doi: 10.1111/j.1574-6941.2011.01153.x
[48] Yanagawa K, Sunamura M, Lever M A, et al. Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the eastern Japan Sea offshore Joetsu [J]. Geomicrobiology Journal, 2011, 28(2): 118-129. doi: 10.1080/01490451003709334
[49] Chen Y, Xu C L, Wu N Y, et al. Diversity of anaerobic methane oxidizers in the cold seep sediments of the Okinawa Trough [J]. Frontiers in Microbiology, 2022, 13: 819187. doi: 10.3389/fmicb.2022.819187
[50] Niu M Y, Fan X B, Zhuang G C, et al. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea [J]. FEMS Microbiology Ecology, 2017, 93(9): fix101.
[51] Lv Y X, Yang S S, Xiao X, et al. Stimulated organic carbon cycling and microbial community shift driven by a simulated cold-seep eruption [J]. mBio, 2022, 13(2): e0008722. doi: 10.1128/mbio.00087-22
[52] Hoehler T M, Alperin M J, Albert D B, et al. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium [J]. Global Biogeochemical Cycles, 1994, 8(4): 451-463. doi: 10.1029/94GB01800
[53] Hallam S J, Putnam N, Preston C M, et al. Reverse methanogenesis: testing the hypothesis with environmental genomics [J]. Science, 2004, 305(5689): 1457-1462. doi: 10.1126/science.1100025
[54] 陈颖. 厌氧甲烷氧化微生物代谢分子机制及其潜在参与矿物形成机理的研究[D]. 上海交通大学博士学位论文, 2014.
CHEN Ying. Molecular metabolism study on microbial anaerobic methane oxidation and the associated biogenic minerals[D]. Doctor Dissertation of Shanghai Jiao Tong University, 2014.
[55] Valentine D L, Reeburgh W S. New perspectives on anaerobic methane oxidation: minireview [J]. Environmental Microbiology, 2000, 2(5): 477-484. doi: 10.1046/j.1462-2920.2000.00135.x
[56] Moran J J, Beal E J, Vrentas J M, et al. Methyl sulfides as intermediates in the anaerobic oxidation of methane [J]. Environmental Microbiology, 2008, 10(1): 162-173.
[57] Raghoebarsing A A, Pol A, van de Pas-Schoonen K T, et al. A microbial consortium couples anaerobic methane oxidation to denitrification [J]. Nature, 2006, 440(7086): 918-921. doi: 10.1038/nature04617
[58] Ettwig K F, Shima S, van de Pas-Schoonen K T, et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea [J]. Environmental Microbiology, 2008, 10(11): 3164-3173. doi: 10.1111/j.1462-2920.2008.01724.x
[59] Chen J, Jiang X W, Gu J D. Existence of novel phylotypes of nitrite-dependent anaerobic methane-oxidizing bacteria in surface and subsurface sediments of the South China Sea [J]. Geomicrobiology Journal, 2015, 32(1): 1-10. doi: 10.1080/01490451.2014.917742
[60] Padilla C C, Bristow L A, Sarode N, et al. NC10 bacteria in marine oxygen minimum zones [J]. The ISME Journal, 2016, 10(8): 2067-2071. doi: 10.1038/ismej.2015.262
[61] 吴忆宁, 梅娟, 沈耀良. 甲烷厌氧氧化机理及其应用研究进展[J]. 生态科学, 2018, 37(4):231-240
WU Yining, MEI Juan, SHEN Yaoliang. Research progress on microbial mechanism and application of anaerobic oxidation of methane [J]. Ecological Science, 2018, 37(4): 231-240.
[62] Hansen L B, Finster K, Fossing H, et al. Anaerobic methane oxidation in sulfate depleted sediments: effects of sulfate and molybdate additions [J]. Aquatic Microbial Ecology, 1998, 14(2): 195-204.
[63] Joye S B, Boetius A, Orcutt B N, et al. The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps [J]. Chemical Geology, 2004, 205(3-4): 219-238. doi: 10.1016/j.chemgeo.2003.12.019
[64] Niemann H, Duarte J, Hensen C, et al. Microbial methane turnover at mud volcanoes of the Gulf of Cadiz [J]. Geochimica et Cosmochimica Acta, 2006, 70(21): 5336-5355. doi: 10.1016/j.gca.2006.08.010
[65] Parkes R J, Cragg B A, Banning N, et al. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark) [J]. Environmental Microbiology, 2007, 9(5): 1146-1161. doi: 10.1111/j.1462-2920.2006.01237.x
[66] Maignien L, Parkes R J, Cragg B, et al. Anaerobic oxidation of methane in hypersaline cold seep sediments [J]. FEMS Microbiology Ecology, 2013, 83(1): 214-231. doi: 10.1111/j.1574-6941.2012.01466.x
[67] Segarra K E A, Comerford C, Slaughter J, et al. Impact of electron acceptor availability on the anaerobic oxidation of methane in coastal freshwater and brackish wetland sediments [J]. Geochimica et Cosmochimica Acta, 2013, 115: 15-30. doi: 10.1016/j.gca.2013.03.029
[68] Ettwig K F, Zhu B L, Speth D, et al. Archaea catalyze iron-dependent anaerobic oxidation of methane [J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(45): 12792-12796. doi: 10.1073/pnas.1609534113
[69] Scheller S, Yu H, Chadwick G L, et al. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction [J]. Science, 2016, 351(6274): 703-707. doi: 10.1126/science.aad7154
[70] Bar-Or I, Elvert M, Eckert W, et al. Iron-coupled anaerobic oxidation of methane performed by a mixed bacterial-archaeal community based on poorly reactive minerals [J]. Environmental Science & Technology, 2017, 51(21): 12293-12301.
[71] Cai C, Leu A O, Xie G J, et al. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction [J]. The ISME Journal, 2018, 12(8): 1929-1939. doi: 10.1038/s41396-018-0109-x
[72] Yan Z, Joshi P, Gorski C A, et al. A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration [J]. Nature Communications, 2018, 9(1): 1642. doi: 10.1038/s41467-018-04097-9
[73] He Q X, Yu L P, Li J B, et al. Electron shuttles enhance anaerobic oxidation of methane coupled to iron(III) reduction [J]. Science of the Total Environment, 2019, 688: 664-672. doi: 10.1016/j.scitotenv.2019.06.299
[74] Liang L W, Wang Y Z, Sivan O, et al. Metal-dependent anaerobic methane oxidation in marine sediment: insights from marine settings and other systems [J]. Science China Life Sciences, 2019, 62(10): 1287-1295. doi: 10.1007/s11427-018-9554-5
[75] He Z F, Zhang Q Y, Feng Y D, et al. Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane [J]. Science of the Total Environment, 2018, 610-611: 759-768. doi: 10.1016/j.scitotenv.2017.08.140
[76] Fu L, Li S W, Ding Z W, et al. Iron reduction in the DAMO/Shewanella oneidensis MR-1 coculture system and the fate of Fe(II) [J]. Water Research, 2016, 88: 808-815. doi: 10.1016/j.watres.2015.11.011
[77] Sivan O, Antler G, Turchyn A V, et al. Iron oxides stimulate sulfate-driven anaerobic methane oxidation in seeps [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(40): E4139-E4147.
[78] Yang H L, Yu S, Lu H L. Iron-coupled anaerobic oxidation of methane in marine sediments: a review [J]. Journal of Marine Science and Engineering, 2021, 9(8): 875. doi: 10.3390/jmse9080875
-



 下载:
下载: